Mining function process links from pathway databases
We can use existing pathway database to mine potential F-P links
For the latest results, see:
Issues
Lack of xrefs in either GO or pathway DB
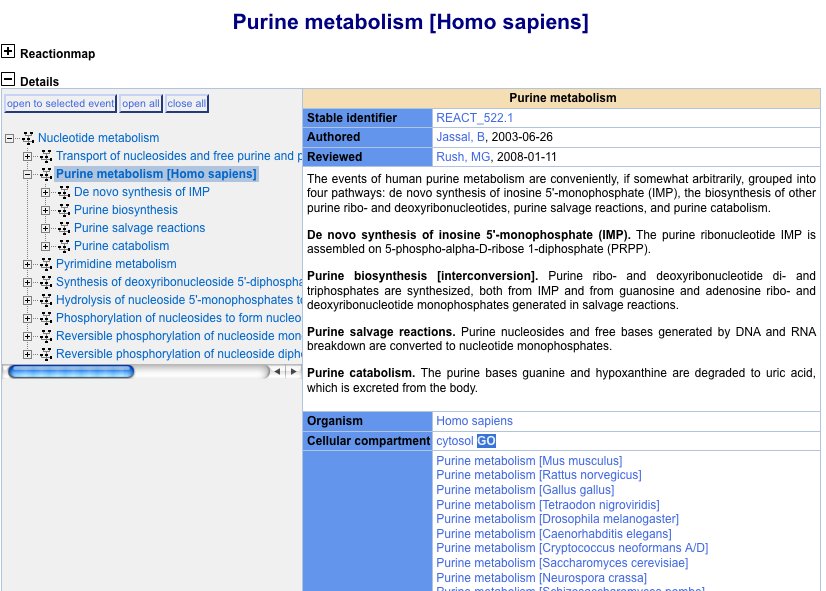
There are many case where there is a pathway ID with no corresponding process ID. We need to do more work to ensure pathways are covered. For example, GO obsoleted "purine metabolism" -- yet this is a pathway in Reactome.
Similarly, Reactome has purine biosynthesis, not present in GO. Instead GO has:
- GO:0006164 ! purine nucleotide biosynthetic process
- GO:0009113 ! purine base biosynthetic process
- GO:0042451 ! purine nucleoside biosynthetic process

|
 http://www.reactome.org/cgi-bin/eventbrowser?DB=gk_current&FOCUS_SPECIES=Homo%20sapiens&ID=73847& http://www.reactome.org/cgi-bin/eventbrowser?DB=gk_current&FOCUS_SPECIES=Homo%20sapiens&ID=73847&
|
Automating detecting xrefs
Some can be found by simple name matching of the GO process and the reaction name:
We can also use XP:biological_process_xp_chebi to mine xrefs.
For example, Formation_of_Acetoacetic_Acid Reactome:77110 currently has no xref to a GO BP (2008/10/13).
However, from the computable pathway description in reactome, we know that the output of this process is CHEBI:15344-acetoacetic acid.
From chebi cross-products we know that GO:0043441-acetoacetic acid biosynthetic process outputs the same type of chemical entity. Thus we can infer an xref between the two.
Inconsistent reciprocal xrefs
see reactome-Homo_sapiens.xrefcheck
Androgen biosynthesis in Reactome xrefs to GO:0006702 ! androgen biosynthetic process
However, there is a lack of reciprocal link:
[Term] id: GO:0006702 name: androgen biosynthetic process namespace: biological_process def: "The chemical reactions and pathways resulting in the formation of androgens, C19 steroid hormones that can stimulate the development of male sexual characteristics." [ISBN:0198506732 "Oxford Dictionary of Biochemistry and Molecular Biology"] synonym: "androgen anabolism" EXACT [] synonym: "androgen biosynthesis" EXACT [] synonym: "androgen formation" EXACT [] synonym: "androgen synthesis" EXACT [] is_a: GO:0006694 ! steroid biosynthetic process is_a: GO:0008209 ! androgen metabolic process is_a: GO:0042446 ! hormone biosynthetic process
Note: in Mining_Process_Function_Links_from_Reactome I use reactome xrefs to GO, not vice versa, so this is not a problem here. However, it is perhaps a problem for people consuming things from the GO side.
Xrefs too general
We have various cases like this:
Reactome:163765 ChREBP activates metabolic gene expression > GO:positive regulation of transcription ; GO:0045941
Imre: The intended semantics is that the goBiologicalProcess slot of a Reactome Event is filled with the equivalent GO biological process term. As evident from your example above this is not always the case - quite often it get's filled with the closest or best fitting (least inappropriate?) GO biological process term. Which is a pity, since we now don't know where we have the clear equivalency and where some sort of approximation.
Peter: No, the intent is that the GO term should be equal in generality to the Reactome event or, if no equal term is available, then a more general GO term should be used. The ChREBP event is not a counterexample, in fact, even though its name makes it look like one. In fact, the process we have annotated is the formation of an active complex of ChREBP with a second protein, MLX, and then the workings of that complex to positively regulate transcription of a number of specific genes
Marc: That would be great, and along with Chris and Imre's suggestions I think that it would be really nice if this autosuggestion feature was available to the curators as they work on modules.
(note: ORB would be useful here)
IMG
In progress