Ontology Structure: Difference between revisions
mNo edit summary |
|||
| (16 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category:Ontology]] [[Category:GO Consortium Meetings]] | |||
Note: this page may get merged with [[Ontology_Development]] | Note: this page may get merged with [[Ontology_Development]] | ||
| Line 4: | Line 5: | ||
DONE | DONE | ||
== ''disjoint_from'' links == | |||
3 terms in GO are no declared mutually disjoint: | |||
CP, MCOP, MOP | |||
Could be done for 3 root nodes | |||
Also for other higher level terms | |||
== Create separate editorial version of ontology == | == Create separate editorial version of ontology == | ||
This page has moved: | |||
[[Ontology_CVS_Directory_Layout_Overhaul]] | |||
== Ontology linking and cross-products == | |||
=== Internal (Intra-GO) links === | |||
GO currently has links within the 3 ontologies, but not between | |||
==== Biological process to molecular function ==== | |||
[[Function-Process_Links]] | |||
==== Cellular component to molecular function ==== | |||
* [ftp://ftp.geneontology.org/pub/go/scratch/obol_results/cellular_component_links_to_molecular_function.obo go/scratch/obol_results/cellular_component_links_to_molecular_function.obo] | |||
==== Implementation ==== | |||
Links spanning the 3 ontologies do not differ significantly from existing DAG links. We could thus include them in the main GO file. However, users expectations will be violated - annotations cannot be propagated up these links in the same way. For this reason it may be better to capture these links in a separate file; for example: | |||
go/ | go/ | ||
ontology/ | ontology/ | ||
gene_ontology_editorial.obo | |||
function_to_process.obo | |||
component_to_function.obo | |||
===== OBO-Edit ===== | |||
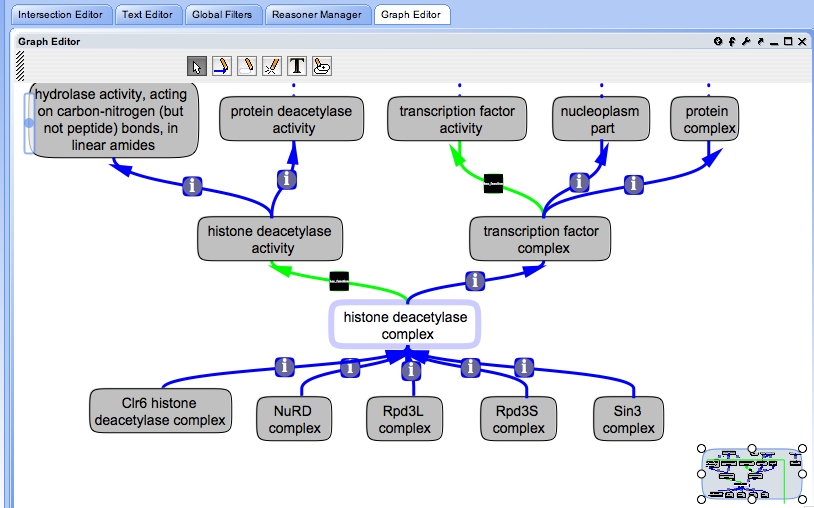
OBO-Edit is capable of handling these links | |||
[[Image:hd-complex.jpg]] | |||
==== | ===== AmiGO ===== | ||
[[AmiGO:Inter-ontology_links]] | |||
=== Internal cross-products === | === Internal cross-products === | ||
| Line 42: | Line 60: | ||
==== Regulation cross-products ==== | ==== Regulation cross-products ==== | ||
[[ | [[Regulation_cross-products]] | ||
==== Other internal cross-products ==== | ==== Other internal cross-products ==== | ||
* [ftp://ftp.geneontology.org/pub/go/scratch/obol_results/biological_process_xp_cell-obol.obo go/scratch/obol_results/biological_process_xp_cell-obol.obo] | |||
=== External DAG links === | === External DAG links === | ||
External DAG links are different from external XPs. A DAG link is just a simple link that happens to span two ontologies. | |||
Examples: between microvillus and hair cell | |||
=== External cross-products === | === External cross-products === | ||
Cross-product meetings: | |||
[http://wiki.geneontology.org/index.php/XP:Meetings] | |||
We have added a [[Cross_Product_Guide]], intended to answer all your questions about cross-products | |||
==== biological_process_xp_cell ==== | |||
CL is currently undergoing rennovation. However, the CL cross-products can still be edited and used. | |||
[[Cell cross-products]] | |||
==== biological_process_xp_chebi ==== | |||
Initial set by mike Bada | |||
[[Chebi cross-products]] | |||
Ongoing issues: | |||
* plurals in ChEBI | |||
* roles in chebi | |||
* unusual DAG structure | |||
==== Others ==== | |||
[http://www.berkeleybop.org/obol Obol] | |||
Latest revision as of 11:54, 14 March 2019
Note: this page may get merged with Ontology_Development
is_a completeness
DONE
disjoint_from links
3 terms in GO are no declared mutually disjoint:
CP, MCOP, MOP
Could be done for 3 root nodes
Also for other higher level terms
Create separate editorial version of ontology
This page has moved: Ontology_CVS_Directory_Layout_Overhaul
Ontology linking and cross-products
Internal (Intra-GO) links
GO currently has links within the 3 ontologies, but not between
Biological process to molecular function
Cellular component to molecular function
Implementation
Links spanning the 3 ontologies do not differ significantly from existing DAG links. We could thus include them in the main GO file. However, users expectations will be violated - annotations cannot be propagated up these links in the same way. For this reason it may be better to capture these links in a separate file; for example:
go/
ontology/
gene_ontology_editorial.obo
function_to_process.obo
component_to_function.obo
OBO-Edit
OBO-Edit is capable of handling these links
AmiGO
Internal cross-products
Regulation cross-products
Other internal cross-products
External DAG links
External DAG links are different from external XPs. A DAG link is just a simple link that happens to span two ontologies.
Examples: between microvillus and hair cell
External cross-products
Cross-product meetings: [1]
We have added a Cross_Product_Guide, intended to answer all your questions about cross-products
biological_process_xp_cell
CL is currently undergoing rennovation. However, the CL cross-products can still be edited and used.
biological_process_xp_chebi
Initial set by mike Bada
Ongoing issues:
- plurals in ChEBI
- roles in chebi
- unusual DAG structure