SO:Composite Terms
SO contains cross-product definitions (aka genus-differentia definitions, aka intersection definitions) for many composite terms. This document describes the methodology. Some familiarity with the obo file format is assumed.
This document is aimed primarily at ontology editors and technical/software/database people who consume the ontologies. It isn't intended for the end-users of ontologies, much of this will be invisible to them.
Pre-crossproducts
Here is an example of a term done using the pre- crossproduct methodology:
[Term] id: SO:0000283 name: engineered_foreign_transposable_element_gene is_a: SO:0000111 ! transposable_element_gene is_a: SO:0000281 ! engineered_foreign_gene is_a: SO:0000805 ! engineered_foreign_region
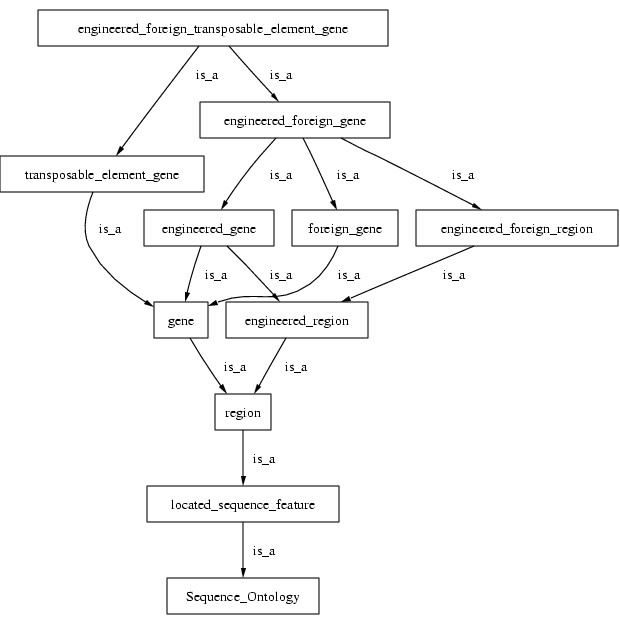
This is problematic. We multiple is_a parents, due to a lack of consistent axis of classification. This leads to tangled DAGs and problems of ontology maintenance, visualisation and reasoning.
Note the editor has to manually check for possible other is_a parents such as "engineered_transposable_elemenent_gene" (ETEG). Furthermore, if ETEG is added, the is_a parentage of EFTEG must be changed. This is tedious, time consuming and error-prone.
The problems continue further up the DAG:
[Term] id: SO:0000281 name: engineered_foreign_gene is_a: SO:0000280 ! engineered_gene is_a: SO:0000285 ! foreign_gene is_a: SO:0000804 ! engineered_region
If we were to examine the whole DAG we would see a lot of redundancy, and no modularisation
Here is an example (showing *is_a* only):
The cross-products solution
The first aspect of the solution is modularity. We realise the separation between the core feature types (such as gene, region) and between the qualities (properties, attributes) of those features. Examples of feature qualities are "being engineered" and "being foreign". These live in a separate part of the ontology, and trace their is_a parentage solely to "feature_attribute", not to "located_sequence_feature".
We also introduce a new relation "has_quality", which obtains between some kind of quality-bearing entity (such as a gene) and a quality.
Using these ingredients we can provide 'Genus-differentia' definitions of terms in a form that is computationally visible. In a definition of this form, a term is defined using a broader category (the genus), and a collection characteristics that distinguish from other instances in the same category (the differentia).
http://en.wikipedia.org/wiki/Definition_by_genus_and_difference
Genus-differentia definitions form one of the core best practices in the OBO Foundry (http://www.obofoundry.org). These definitions can be written as "A <G> 'which' <D>". For example, we can define an engineered foreign transposable element gene as "A transposable element gene *which* is engineered and is foreign". The genus is "tranposable element gene" and the differentia are "is engineered" and "is foreign".
We can also expose these definitions in a way that is computationally visible. [add picture of editing in oboedit here].
obo file representation
The underlying representation in oboedit is as follows:
[Term] id: SO:0000283 name: engineered_foreign_transposable_element_gene intersection_of: SO:0000111 ! transposable_element_gene intersection_of: has_quality SO:0000783 ! engineered intersection_of: has_quality SO:0000784 ! foreign
The "intersection_of" lines list the necessary and sufficient conditions for inclusion in a class (term). For this to be a G-D definition, there should be one intersection_of line without a relation (the genus) and at least one line with a relation (the differentia).
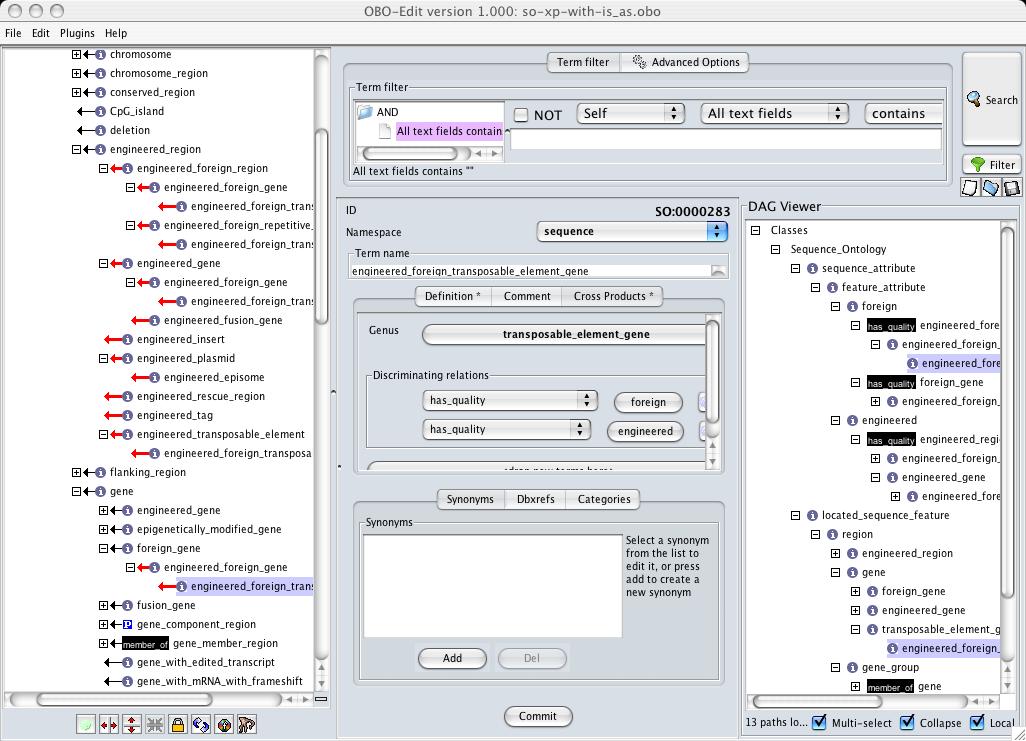
Of course, most people will not be looking at obo files. Oboedit provides a plugin for editing these genus-differentia definitions (see below for screenshot)
Using these definitions, a computer can calculate where EFTEG should be placed in a DAG (provided similar definitions are provided for other terms). The computer can also calculate that EFTEGs should be returned in queries for ETEGs or EFRs (engineered_foreign_regions).
These caclulations are typically done with a 'reasoner'. oboedit has a reasoner built-in.
The blue squiggly lines are 'is_a's that have been inferred by oboedit using the genus-differentia definitions. They have 'not' been asserted by the person editing the ontology.
This is all well and good for oboedit users, but not everyone uses uses this tool. Whilst there are many other reasoners available, we should still provide the DAG fully classified so that there are no additional dependencies required by consumers of the ontology.
We can configure oboedit to save all inferred 'is_a' links (see issues, below). The saved file will have entries like this:
[Term] id: SO:0000283 name: engineered_foreign_transposable_element_gene intersection_of: SO:0000111 ! transposable_element_gene intersection_of: has_quality SO:0000783 ! engineered intersection_of: has_quality SO:0000784 ! foreign is_a: SO:0000111 ! transposable_element_gene is_a: SO:0000281 ! engineered_foreign_gene
We call the is_a links above 'asserted', because they are explicitly stated in the file, rather than implicitly inferred by the oboedit reasoner.
This means that software can ignore the intersection_of lines safely, the old tangled DAG can still be displayed as normal.
When the ontology with asserted 'is_a' links is viewed in oboedit, it will look like this:
The red arrows indicate asserted 'is_a' links that could have been inferred had they not been there
Obtaining
The public version of the ontology contains the logical definitions
The genus-differentia matrix can be manipulated as an excel file
Media:so-xp.xls -- generated 2006/08/25
Benefits
The management of the tangled is_a DAG is handled automatically by software, so the ontology editor does not need to worry about it. Downstream tools should not be affected.
However, second-generation tools can choose to use the intersection_of lines; they can be used to present the ontology DAG to the user in a more tractable, modular fashion. The genus in the definition can be used as the "core" is_a parent. The differentia could be presented in a separate display.
open issues
saving inferences
oboedit does not allow you to save all inferred 'is_a's. Currently so-xp is saved without the inferred is_a parents which limits its applicability to first-generation obo tools (ie those without reasoning capabilities).
Until oboedit can do this, it may be necessary to semi-manually add the is_as (oboedit shows you these visually but it doesn't provide a way to materialize them in the resulting saved obo file).
Another option is to convert to owl and use a third-party open source reasoner such as pellet to do the classification, then convert back to obo. This could all be automated in a script. The curator version (so-xp.obo) would not have the is_as, but the so.obo file that is for public consumption and use by first-generation tools would have the is_as materialised.
UPDATE: we used Pellet to do the initial classification. Results still being checked. Once John is back we can discuss ways of making it easier to save the oboedit classification results, or using obo2obo to fill these in, but Pellet seemed to work as a one-off
http://www.mindswap.org/2003/pellet/
what happens on changes?
One advantage in never asserting the inferrable 'is_a' links is never having to worry about recreating 'is_a links when the core parts of the ontology change.
For example, if we were to create an intermediate type between "gene" and "region" (for example, "functional region") and also wanted to created terms like "engineered functional region") we would simply go ahead and do that, provide genus-differentia definitions, and let the reasoner compute the is_a DAG on-the-fly.
However, as we stated earlier, we want to save the obo file with the DAG fully classified, since most tools that consume the obo file will not be reasoner-aware. We can still use oboedit to create the is_a links automatically, and configure it so that these are saved. The problem here is that change in one part of the ontology can percolate to large sections of the DAG - how do we know which links to replace and which to preserve?
One way is to keep around information on which links were asserted directly by a curator not as a result of reasoning, and which were originally asserted by the reasoner? For example, we could use trailing qualifiers:
[Term]
id: SO:0000283
name: engineered_foreign_transposable_element_gene
intersection_of: SO:0000111 ! transposable_element_gene
intersection_of: has_quality SO:0000783 ! engineered
intersection_of: has_quality SO:0000784 ! foreign
is_a: SO:0000111 ! transposable_element_gene {inferred=true}
is_a: SO:0000281 ! engineered_foreign_gene {inferred=true}
The reasoner would know that these could be discarded if they can no longer be inferred.
This is still under discussion. For now, these links may have to be removed manually - which is no worse than the pre-reasoner situation when everything was done manually
Re-Use
Currently SO has its own ontology of feature attributes; eventually we may want to merge this with PATO PATO:Main_Page
So also uses its own has_quality relation. Eventually it should use the version that will be in RO RO:Main_Page.
applicability of methodology to other ontologies
This work was carried out as part of a larger project within the Gene Ontology and the http://www.obofoundry.org [OBO-Foundry] to create logical and computable genus-differentia definitions for terms, linking across ontologies where appropriate. See XP:Main_Page
We are applying the same methodology to GO, although the xps are not yet part of the public release. We are focused on xps for GO terms that refer to CL terms right now.
other resources
mail lists
https://lists.sourceforge.net/lists/listinfo/obo-crossproducts
oboedit guide
Link to appropriate section of oboedit guide here...
background reading
definitions in the OBO Foundry
Forthcoming paper
Obol paper; see link on: http://www.fruitfly.org/~cjm/obol
Modularity in ontologies
These tutorials are very OWL and Protege centric, but much of it also applies to obo1.2 and oboedit: