Regulation cross-products: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
For more background, see this presentation: | |||
* [[Media:go-sw-report-2006-cambridge.ppt]] | |||
* | |||
==Regulation cross-products== | |||
The oboedit reasoner can be used to automatically manage regulation terms; this can serve as a trial run before implementing cross-products with external ontologies. | The oboedit reasoner can be used to automatically manage regulation terms; this can serve as a trial run before implementing cross-products with external ontologies. | ||
| Line 12: | Line 9: | ||
For now we mainly consider internal regulation terms; ie regulation of biological process. | For now we mainly consider internal regulation terms; ie regulation of biological process. | ||
You will need the file | You will need the file gene_ontology_xp.obo, from: | ||
ftp://ftp.geneontology.org/pub/go/scratch | |||
* ftp://ftp.geneontology.org/pub/go/scratch | |||
(this file should be updated daily from | (this file should be updated daily from gene_ontology_edit.obo, provided my cron works) | ||
You can load this into oboedit. Turn the reasoner on; then read on for an explanation | You can load this into oboedit. Turn the reasoner on; then read on for an explanation | ||
| Line 32: | Line 30: | ||
# things that regulate synpatogenesis | # things that regulate synpatogenesis | ||
(This also involves introducing a new relation, '''regulates''') | |||
Here is what this looks like in oboformat: | Here is what this looks like in oboformat: | ||
| Line 44: | Line 42: | ||
is_a: GO:0051963 ! regulation of synaptogenesis | is_a: GO:0051963 ! regulation of synaptogenesis | ||
intersection_of: GO:0048519 ! negative regulation of biological process | intersection_of: GO:0048519 ! negative regulation of biological process | ||
intersection_of: | intersection_of: regulates GO:0007416 ! synaptogenesis | ||
| Line 59: | Line 57: | ||
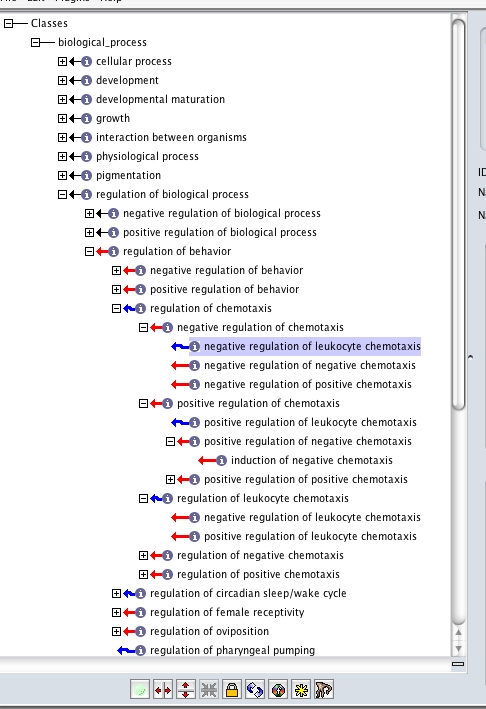
[[Image:Regulation-with-reasoner.jpg]] | [[Image:Regulation-with-reasoner.jpg]] | ||
(See the presentation above for more up-to-date screenshots) | |||
here oboedit is saying that '''-reg of chemotaxis''' ''can be inferred to be an is_a child of'' '''-reg of behavior''' | here oboedit is saying that '''-reg of chemotaxis''' ''can be inferred to be an is_a child of'' '''-reg of behavior''' | ||
| Line 66: | Line 66: | ||
Or you can use obo2obo; like this: | Or you can use obo2obo; like this: | ||
obo2obo | obo2obo gene_ontology_xp.obo -o -saveimpliedlinks -realizeimpliedlinks gene_ontology_xp-with-implied-links-saved.obo | ||
(this step is already done; see | (this step is already done; see the scratch directory) | ||
Here is another example: | Here is another example: | ||
| Line 89: | Line 89: | ||
genus: {-/+} regulation of biological process | genus: {-/+} regulation of biological process | ||
differentia: | differentia: regulates X | ||
in obo format this is | in obo format this is | ||
intersection_of: GO:ID_for_{-/+}_regulation_of_BP | intersection_of: GO:ID_for_{-/+}_regulation_of_BP | ||
intersection_of: | intersection_of: regulates GO:ID_for_X | ||
the oboedit reasoner does the rest | the oboedit reasoner does the rest | ||
| Line 109: | Line 109: | ||
Eventually it would be good to have an excel style table for xp editing in oboedit; imagine a table with 3 rows (reg, +reg, -reg) and a columns for core processes (perhaps limited to a branch of the DAG); each cell filled in with the ID of the xp term. New terms added simply by clicking on the empty cell; names, synonyms and defs populated automatically. | Eventually it would be good to have an excel style table for xp editing in oboedit; imagine a table with 3 rows (reg, +reg, -reg) and a columns for core processes (perhaps limited to a branch of the DAG); each cell filled in with the ID of the xp term. New terms added simply by clicking on the empty cell; names, synonyms and defs populated automatically. | ||
===Related=== | |||
In the directory | |||
* ftp://ftp.geneontology.org/pub/go/scratch | |||
You will also see the report for the disjointness violations in the DAG. See the presentation above for an example | |||
Revision as of 22:45, 20 January 2007
For more background, see this presentation:
Regulation cross-products
The oboedit reasoner can be used to automatically manage regulation terms; this can serve as a trial run before implementing cross-products with external ontologies.
For now we mainly consider internal regulation terms; ie regulation of biological process.
You will need the file gene_ontology_xp.obo, from:
(this file should be updated daily from gene_ontology_edit.obo, provided my cron works)
You can load this into oboedit. Turn the reasoner on; then read on for an explanation
Logical (cross-product) Definitions
oboedit needs the definition (necessary and sufficient conditions) of regulation terms to be made explicit rather than buried in text. We do this using the new oboformat1.2 intersection_of (cross-product definition) tag.
The idea is that we define a term like negative regulation of synaptogenesis as being:
- A negative regulation process in which the the process being regulated is synpatogenesis
This is an aristotelian (genus-differentia) definition. It can also be seen as the cross-product (intersection) of:
- negative regulation of biological process
- things that regulate synpatogenesis
(This also involves introducing a new relation, regulates)
Here is what this looks like in oboformat:
[Term] id: GO:0051964 name: negative regulation of synaptogenesis namespace: biological_process def: "Any process that stops, prevents or reduces the frequency, rate or extent of synaptogenesis, the formation of a synapse." [] is_a: GO:0051961 ! negative regulation of nervous system development is_a: GO:0051963 ! regulation of synaptogenesis intersection_of: GO:0048519 ! negative regulation of biological process intersection_of: regulates GO:0007416 ! synaptogenesis
Here we have added two lines. These lines can be safely ignored by obof1.2 unaware parsers; they can be stripped out prior to making public if need be. However, they provide oboedit (or any other reasoner-aware tool, like Protege/SWOOP) with the information required to automatically manage the placement of these terms in the DAG.
In the oboedit cross-product box, this should look like:
Genus: negative regulation of biological process Differentia: part_of synaptogenesis
Using the reasoner
open the file in the reasoner, you should be able to see both missing is_a links (blue squiggly lines) as well as redundant links (straight red lines).
(See the presentation above for more up-to-date screenshots)
here oboedit is saying that -reg of chemotaxis can be inferred to be an is_a child of -reg of behavior
oboedit can infer this from the logical definition (intersection_of lines). Note that the actual is_a links are not there in the underlying obo file - this is the inferred (implied) graph, not the asserted graph (to see the asserted graph, simply turn off the reasoner; you will see -reg of chemotaxis disappear as a child of -reg of behavior. You can save the inferred graph as an asserted graph (ie write out all the inferred is_a links) using a special save option (JOHN - HOW?).
Or you can use obo2obo; like this:
obo2obo gene_ontology_xp.obo -o -saveimpliedlinks -realizeimpliedlinks gene_ontology_xp-with-implied-links-saved.obo
(this step is already done; see the scratch directory)
Here is another example:
you should be able to see the cross-product definition of the focused term. This term is not asserted to be a child of regulation of cellular process, but this is implied (see the DAG view on the right)
Don't spend too long fixing these yet - this is a trial run. This is too reactive a process to incorporate into production; you want new regulation terms to go straight into the DAG. The next step will be to actually maintain the logical definitions in the gene_ontology_edit.obo file (but strip them from the public one since they are obof1.2). I'll send details later in the week. The basic idea is you will add regulation terms providing the minimal information and oboedit will do everything else.
Note that for now I've simply used the part_of relation in the logical definition - we can discuss whether this is a good time to move to regulates.
How does this work
How does this all work? I have a simplified version of obol that takes a term
{-/+} regulation of X
And creates a logical definition
genus: {-/+} regulation of biological process
differentia: regulates X
in obo format this is
intersection_of: GO:ID_for_{-/+}_regulation_of_BP
intersection_of: regulates GO:ID_for_X
the oboedit reasoner does the rest
Next steps
Currently this is an experimental trial which should hopefully help the IsaComplete process.
The next step could be to place the intersection_of lines directly in the main IsaComplete.obo file, rather than the separate IsaComplete-xp.obo file.
We could then move these into gene_ontology_edit.obo; we will need to check that the conversion process to gene_ontology.obo works fine.
Once the genus-differentia defs move to the live file, it will become best practice to maintain the genus-differentia links directly in the file (though an obol process could help check for errors). This may seem like extra work but in theory it should amount to a lot less. Adding a new regulation term (or 3) should simply be a matter of clicking on the core process term and making a few clicks. oboedit should do the rest. More epxloratory work required here.
Eventually it would be good to have an excel style table for xp editing in oboedit; imagine a table with 3 rows (reg, +reg, -reg) and a columns for core processes (perhaps limited to a branch of the DAG); each cell filled in with the ID of the xp term. New terms added simply by clicking on the empty cell; names, synonyms and defs populated automatically.
Related
In the directory
You will also see the report for the disjointness violations in the DAG. See the presentation above for an example